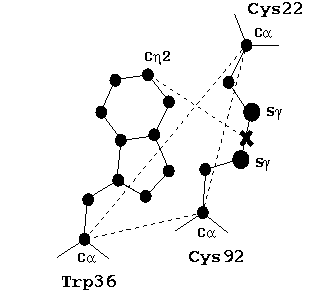

Within the core of the Ig, there is a highly-buried disulfide bridge that connects the two beta-sheets. This bridge has been shown to be one of the most highly conserved parts of fold (Gerstein and Altman, 1995), and has been dubbed the "pin" (Chothia and Lesk, 1982). The next most conserved residue is a tryptophan (found in over 99% of all Ig sequences) that packs over the disulfide bridge. We have recently done a study of the conserved geometry of the relationship between this tryptophan and the disulfide using 60 experimentally-determined high-resolution Ig structures. The configuration of this triad of residues (Cys-Cys-Trp) is extraordinarily conserved. We suspect that this conservation reflects an important role that the tryprophan plays, possibly in protecting the disulfide bond from being reduced in hostile extracellular environments.
Based on our calculations of the canonical geometry of the triad in Ig, we wrote a program to search for similar motifs in other proteins. Basically, the program reads in all of the ATOM records from a PDB file, searches for a pair of cysteines whose C-alpha atoms are within a reasonable distance, and whose S-gamma atoms are within a reasonable distance, and then searches for a tryptophan whose C-alpha atom matches the triad-defined distances to the cysteine C-alpha's, and whose Cn2 atom is close to the midpoint of S-gamma's. For any combination of residues that satisfies these constraints, an analysis is printed out that identifies the residues, gives the distances, and also prints some other useful information (e.g. angles).
Using this program, we found the triad in many other members of the immunoglobulin super-family, including T-cell receptors, class I and class II MHC, CD4, CD8, vascular cell adhesion molecule (VCAM-1), and myelin membrane adhesion molecule (PO). By searching the PDBSelect (a representative set of >600 structures in the the PDB with <25% homology; Hobohm and Sander, 1994), we also found putative triads in several non-Ig proteins, such as galactose oxidase (1gof) and a virus envelope protein (1svb). Note, given knowledge of the parameters of the triad, one could also use other more general programs for searching for motifs in proteins structures (Wallace et al., 1997; Russell, 1998).
Reference:
T.R. Ioerger and D.S. Linthicum (1999). T-SEARCH: A program to identify and measure the geometry of cystein-cysteine tryptophan (C-CW) triad motifs in immunoglobulin superfamily structures and other proteins. Biotechnology Software and Internet Journal, 16(3):26-31.
The program is run from the command line and takes 1 argument: the name of a PDB file. It reads the PDB and prints out information on triads it find. The program has negligible runtime, e.g. ~1 sec on an O2. You may also run the program with the flag "-c", which prints out the constraints used in the search.
Source code:
Pre-compiled binaries: Notes:
Here are the constraints currently used to recognize triads:
unix> triads -c *************************************************************** * TRIADS * * Thomas R. Ioerger, copyright 1998 * * http://www.cs.tamu.edu/faculty/ioerger/triads/triads.html * *************************************************************** triad constraints... atom 1 atom 2 typical traid dist max dist cutoff Cys1-CA Cys2-CA 6.5 A 8 A Cys1-SG Cys2-SG 2.0 A 3 A Cys1-CA Trp-CA 4.5 A 6 A Cys2-CA Trp-CA 9.5 A 12 A Trp-Cn2 S-S midp. 4.5 A 7 A
Here is some example output from 1baf, which is an immunoglobulin:
unix> triads 1baf.pdb *************************************************************** * TRIADS * * Thomas R. Ioerger, copyright 1998 * * http://www.cs.tamu.edu/faculty/ioerger/triads/triads.html * *************************************************************** triad: 1: CYS-87L-CA (798): <-13.658,48.203,180.111> 2: CYS-23L-CA (194): <-19.514,48.748,182.37> 3: TRP-34L-CA (298): <-12.765,50.081,176.074> dist(CA1,CA2)=6.30023 dist(CA1,CA3)=4.5411 dist(CA2,CA3)=9.32552 dist(Trp-Ch2,SS-midpoint)=4.78824 angle between Cys C-alpha/C-beta vectors = 144.439 degrees dihedral angle across S-S bond = 73.8296 degrees triad: 1: CYS-194L-CA (1828): <-19.705,7.407,173.404> 2: CYS-134L-CA (1215): <-14.788,10.92,170.976> 3: TRP-148L-CA (1361): <-19.464,7.302,178.531> dist(CA1,CA2)=6.51255 dist(CA1,CA3)=5.13373 dist(CA2,CA3)=9.59339 dist(Trp-Ch2,SS-midpoint)=4.93975 angle between Cys C-alpha/C-beta vectors = 163.578 degrees dihedral angle across S-S bond = -95.5199 degrees triad: 1: CYS-96H-CA (2971): <4.278,45.653,177.569> 2: CYS-22H-CA (2223): <10.311,44.681,176.239> 3: TRP-37H-CA (2365): <3.846,46.256,181.861> dist(CA1,CA2)=6.25386 dist(CA1,CA3)=4.35562 dist(CA2,CA3)=8.71112 dist(Trp-Ch2,SS-midpoint)=4.69424 angle between Cys C-alpha/C-beta vectors = 132.524 degrees dihedral angle across S-S bond = 68.6673 degrees triad: 1: CYS-197H-CA (3861): <-0.648,15.395,160.883> 2: CYS-142H-CA (3368): <-4.265,13.03,166.113> 3: TRP-156H-CA (3494): <-2.624,18.772,158.457> dist(CA1,CA2)=6.78446 dist(CA1,CA3)=4.60371 dist(CA2,CA3)=9.70968 dist(Trp-Ch2,SS-midpoint)=3.99427 angle between Cys C-alpha/C-beta vectors = 140.278 degrees dihedral angle across S-S bond = -90.2902 degrees